siRNA Screening Overview
Introduction
This report is designed to be an introduction to High Throughput Screening (HTS) at the Target Discovery Institute, Nuffield Department of Medicine, University of Oxford. Below you will find information detailing all aspects of HTS performed at our facility including examples of completed screens and their results. A broad range of screens have been run in our facility and our scientists have extensive knowledge of high throughput screening. We are available for consultation at the earliest stages of assay conception and we will work closely with each investigator through all stages of assay development, optimization, automation and screening.
HTS Transfer Protocols
All assays being transferred to the Janus for siRNA HTS will be subjected to a thorough validation process before the actual screen assay (production) proceeds.
This validation process will consist of:
1. Initial Consultation
2. Stability and Process study
3. Liquid Handling Validation
4. Plate Uniformity Assessment
5. Replicate Experiment
6. Production Runs
1. Initial Consultation
The initial consultation with the HTS facility manager is an opportunity to discuss your ideas for a potential screen during the conceptual stage or as you begin the bench-top development. We have run a wide variety of screens in our facility and have a broad knowledge of what types of screens might be most effective (time and cost) for the question you would like to answer. We have also been successful in developing novel screens designed to answer specific questions and would welcome the opportunity to develop additional novel screens. We would encourage you to discuss your proposed assay at the earliest stages of development as we can assist you with the following issues:
- 1. There are several types and many approaches to HTS. Each type of screen has merits and demerits (i.e. - easy to set up but requires more consumables).
- 2. Your choice of reagents - Some reagents have limited stability, may be very expensive compared to alternatives, susceptible to pH or temperature changes, etc. make them unsuitable for HTS.
- 3. Transfect ion Optimization - Optimizing the transfection efficiency is the single most crucial step to successful RNAi screens. We can help you devise a specific transfection optimization protocol to target the best cell density, transfection reagent, target gene knock-down, and cytotoxicity.
- 4. We can begin to discuss logistics and timing of your assay as well as the projected cost.
An early initial consultation will ensure a smooth transition from bench-top to HTS.
2. Stability and Process
The stability and process study is the first step of the HTS assay validation. It will be necessary to examine all reagents used in the assay for the following properties:
- 1. Stability during production.
- 2. Storage stability (For unused reagents you may need to save).
- 3. Emergency stability (the stability of assay reagents at room temperature in the unlikely event of a robotics failure).
- 4. Time-course experiments to determine the range of acceptable times for each reagent in process.
This study can run concurrently with the final steps of the liquid handling validation and will be the responsibility of assay developer.
3. Liquid Handling Validation
Once your proposed screen assay has met the criteria for passage from the bench-top to the Janus Liquid Handling Work Station, the programming of all assay steps on the liquid handling Robots will become the responsibility of the high throughput developer.
- 1. Initially, we will meet to thoroughly discuss the protocol and any questions or clarification should be addressed here.
- 2. Within a couple of days I will provide you with a written time-line for the development of your screen.
- 3. Once the High Throughput liquid handling protocol is complete, we will validate all steps of the protocol together using colored dyes to track all liquid handling steps. You will thoroughly evaluate the protocol at this point. There will invariably be alterations from your protocol and any variations will be approved by you.
4. Plate Uniformity Assessment
Once your proposed screen assay has passed the Liquid Handling Validation step, a Plate Uniformity Assessment will be conducted (Detailed below in this report). This is a series of experiments designed to address:
- 1. Drift
- 2. Edge Effect
* In general, Drift or Edge Effects <20% are considered acceptable and random effects seen on <20% are also considered acceptable. If HTS protocol adjustments are necessary to address unacceptable Drift or Edge Effects, they will be made by the HTS developer and approved by you.
5. Replicate Experiment - Final Assay Validation
This is a set of experiments designed to validate the HTS procedure immediately before the Production runs. Along with the Plate Uniformity Assessment, the Replicate Experiment will ensure:
- 1. For RNAi screens, a two step transfection optimization procedure using Janus Liquid Handling robots (detailed below in this report) will be conducted at this stage to ensure the transfection conditions are optimized for your screen.
- 2. A second set of experiments, a "dry run", using all components of the HTS assay will be performed. We will seed plates and carry out all steps of the assay to produce a Z' Factor[1], to determine if the screen produces data with sufficient sensitivity to accurately differentiate between positive "hits" and negative samples.
- 3. Plate Variability and Plate-to-Plate Variability will be assessed at this point.
- 4. All Plate and Assay controls will be assessed.
If all aspects of the HTS assay are performing as expected, we will proceed to the Production run.
6. Production Run
- 1. Run the HTS assay. This is where your HTS assay produces reams of data to be written up in high impact scientific journals!
- 2. Post-Production evaluation and report. See appendix 1 for HTS Final Report layout.
Types of High Throughput Screens
High throughput screening assays can be divided into two main types:
1. Protein based biochemical screens - These screens use purified proteins, substrates and small compound inhibitors in buffered solutions to produce an optical (fluorescent, luminescent, etc.) readout that monitors enzymatic or binding activity using high throughput platereaders. Small compound inhibitors are ranked on their ability to reduce the protein function, and by extension the optical signal, in this type of screen.
Advantage: Very high throughput, small volume reactions reduce reagent costs and simple readouts. Target of the inhibitor is defined.
Disadvantage: Small compound may not be water soluble, membrane permeable and may be promiscuous or toxic. Single protein inhibition may not cause desired phenotypic change in cells or tissues due to redundant pathways.
2. Cell based screens- These screens can be RNAi or small compound screens that utilize cells plated in 96 or 384 well plates to produce a visual phenotypic change in the cells which can be measured. The three general measurement types (although others exist) of a cell based screen are:
- 1. Uniform well readouts - These include cell viability assays. These assays usually employ high throughput platereaders to produce their measurements.
- 2. High-Content Imaging Screens - These include small compound as well as RNAi screens and are designed to probe changes to a cellular phenotype (i.e. foci formation screens, nuclear and cellular morphology, localization of proteins, etc). HCI screens employ specialized high content imagers to produce high content pictures which can be used to measure phenotypic changes. These assays are content rich and can provide in-depth analysis of cell images. In addition, they can also be "multi-plexed" with multiple probes and DAPI for DNA staining to provide information on several changes to a cells phenotype simultaneously. They can also provide cell cycle information.
- 3. Reporter gene systems - These are mostly high throughput FACS based assays. They employ high throughput FACS to produce readouts of GFP, luciferase, etc. internalized signal.
Advantages: Cell based assays produce phenotypic changes affecting pathways directly associated with disease states and the target protein does not have to be known. Small compound cell based screens ensure compound solubility, membrane permeability, non-toxic and effectiveness at low therapeutically relevant concentrations.
Disadvantages: Lower throughput than protein based assays, much more technically difficult and much longer duration (days/weeks vs. minutes). Target of inhibitors are not definitively known.
RNAi Screens
The objective of RNAi screens run in the Target Discovery Institute are focused on "target discovery" identifying novel proteins involved in disease pathways, defining the majority of proteins involved in specific repair pathways (i.e. - homologous recombination) and identifying novel synthetic lethal interactions. Additionally, we are focused on identifying proteins that could be used as targets for therapeutic treatments with small compound inhibitors (i.e. - identifying proteins that when inhibited, radio-sensitize cells).
Types of RNAi Screens
Several types of RNAi screens have been employed in the TDI including:
- Loss of Function screens (LOF) - A LOF screen describes a simple phenotypic change (foci formation) or decrease in cell viability when a single gene is knocked-down in an assay. These types of screens can measure the increase in foci formation after treatment with a compound or damage inducing IR. LOF screens are usually the most straight forward type of RNAi screen to run.
It is important to remember that a LOF screen can provide two types of phenotypic change. Either an enhancement of a phenotype (e.g. increased number of foci when compared to positive control) or a decrease or loss of the phenotype (e.g. decreased number of foci when compared to positive control). Both can be very import.
- Synthetic Lethal screen - A synthetic lethal screen employs dual knock-downs or parallel KO cell lines to create a synthetic lethality (when the knock-down of either gene is harmless but the knock-down of two genes in concert causes cell death). Synthetic lethal screens are more difficult because two genes must be knocked-down with high efficiency to observe significant cell death.
- Mini-clonogenic RNAi screen- Clonogenic assays study the effectiveness of drugs or IR on proliferating tumour cells but are expensive and laborious to run on the bench-top. The mini-clonogenic assay is a type of LOF screen where the clonogenic assay has been miniaturized to 96 Well plates.
Above are examples of RNAi screens we have produced at TDI. There of course are several addition types of RNAi screens available to researchers which you may be interested in exploring. We are available for consultation for any screens you may be interested and are well equipped to process these screens in our facility.
Criteria for developing robust RNAi HTS
The HTS conditions should be optimized to satisfy several criteria:
- 1. Since RNAi screens are dependent on efficient transfection, optimization of transfection conditions is crucial to the success of HTS! See the Two Step Janus Transfect ion Protocol below for information on how we optimize transfection conditions in TDI.
- 2. Use only Healthy and robust cells - self explanatory.
- 3. Transfect ion of your cell line with negative control non-targeting (NC-NT) siRNA should not reduce cell viability in excess of 10 to 20% when compared to cells with transfection treatment without siRNA. Cell viability below 20% would indicate no significant siRNA non-specific toxicity.
- 4. The knock-down should produce a robust phenotypic change with a measurable Z' Factor above 0.5 for the control samples. No screen will be progress to production without a robust dynamic range (see below).
- 5. Cast a wide net during the primary screen - establish the optimized transfection conditions but then use 2x the transfection reagent to ensure knock-down. The goal of the primary screen is maximal sensitivity. At this stage, false positives (FPs) are much better than false negatives (FNs)!
- 6. Knock-down does not always equal significant phenotypic change - some genes require >90% knock-down over 96 hours to observe significant phenotypic change.
To help ensure all criteria are satisfied during the development of an HTS and before production runs commence, we employ a two step assay validation protocol using the PerkinElmer Janus liquid handling workstation. The assay validation protocols are detailed below.
Plate Layout and Controls for RNAi Screens
A number of different controls (both positive and negative) are necessary to obtain meaningful and reliable results from RNAi screening.
They are summarized below:
Positive Controls
PC-S This is the positive control for silencing. The PC-Ss are siRNAs that induce a high level of gene knockdown, they are NOT involved in the pathway you are studying and should not target genes that affect cell proliferation or survival (e.g., GAPDH or beta-actin). The PC-S will simply provide information on the efficiency of the positive knockdown in the screen and will NOT be used in the statistical analysis of your data.
PC-A: This is the positive control for your assay. The PC-As are siRNAs that should induce your screening phenotype and CAN BE used in the statistical analysis of your data for evaluating hits. The PC-As should target known genes in your pathway and it is very important to test several potential PC-As to find one that produces the desired phenotypic change at the levels you require.
PC-A2: It is often very helpful if a second PC-A (we'll call it PC-A2) is used that induces a moderate phenotypic change in the assay. This control will not be used in the statistical analysis of the data but can be used as a phenotypic marker to evaluate the results.
Negative Controls
NC-NT: This is the Non-targeting negative control and establishes the baseline for your assay. The NC-NT measures the changes siRNA delivery can make on gene expression. In most cases, this should be a nonsense sequence with no complementary to known genes and should have no effect on your assay results.
NC-NsiRNA: This is the nontransfected negative control and contains only seeded cells with no transfection reagent/siRNAs. The NC-NsiRNA can be used in conjunction with the NC-NT to determine if siRNA delivery affects assay results.
NC-T: This is the treated negative control and is only used in experiments having additional treatments (drugs, chemicals, etc.). The NC-T serves as the baseline for effects of treatment alone on cells.
NC-NC: This is the no-cell negative control. The assay wells are treated with all reagents used during the experiment and measures non-specific signals from these reagents. This control is considered unnecessary for most screens.
Typical plate layout for RNAi screens at TDI
The primary screens in a 96-well and 384-well format should have 16-24 negative wells per plate for the negative control to be used as the negative reference for selecting strong to moderate strength phenotypic assay response. [2]Both the False Negative Rate (FNR) and the False Positive Rate (FPR) are lowest at 24 wells. However, a good trade-off between cost and benefit can be found at sample sizes between 4-11 samples. Since the aim of many primary screens is to capture siRNA knock-down with strong to moderate strength, sample negative control size employed by our primary screens will be between 8-11 samples.
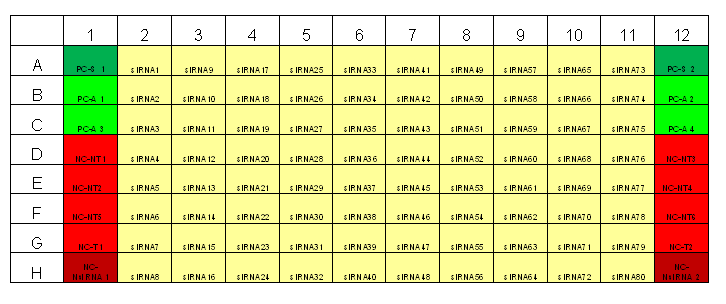
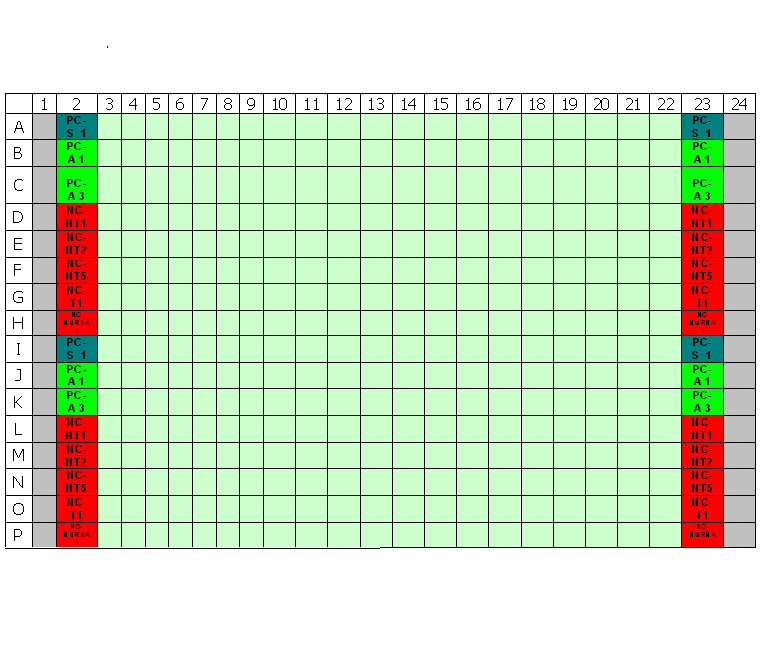
Transfection Optimization using Janus Liquid Handling Robots (Two Step Optimization):
Optimizing the transfection efficiency is the single most crucial step to successful RNAi screens. Below is a protocol for transfection optimization using Janus Liquid Handling Robots (Two Step Optimization), from which we choose the best transfection conditions for our RNAi screens.
Step 1: Set the siRNA and transfection reagent concentrations using the Janus liquid handling robots to produce a matrix of siRNA and transfection reagents. Using two different siRNAs (generally against two proteins with a proven knock down and with defined antibodies for performing Westerns) and two different transfection reagents (chosen through initial studies or experimental experience narrow the TRs available) to produce the matrix. A literature search can also help identify effective transfection reagents. This experiment will establish two criteria:
- 1. At 72 and 96 hours post transfection, a cell viability assay using resazurin is performed to determine transfection reagent toxicity. All transfection reagents are toxic to cells at varying degrees. The toxicity of the NC-NT when compared to cells alone will produce a dose dependant range of toxicity. Transfect ion of your cell line with NC-NT siRNA should not reduce cell viability in excess 20% when compared to cells with transfection treatment without NC-NT and any transfection reagent concentrations that exceed these limits are eliminated from further evaluation.
- 2. Next, the siRNA and transfection reagent concentrations that produce the best knock-down of the proteins are evaluated by harvesting the proteins and running westerns. All conditions are evaluated and the analysis will provide the:
- A. siRNA concentration.
- B. Transfect ion reagent vendor (in conjunction with the transfection reagent viability data).
- C. Transfect ion reagent concentration.
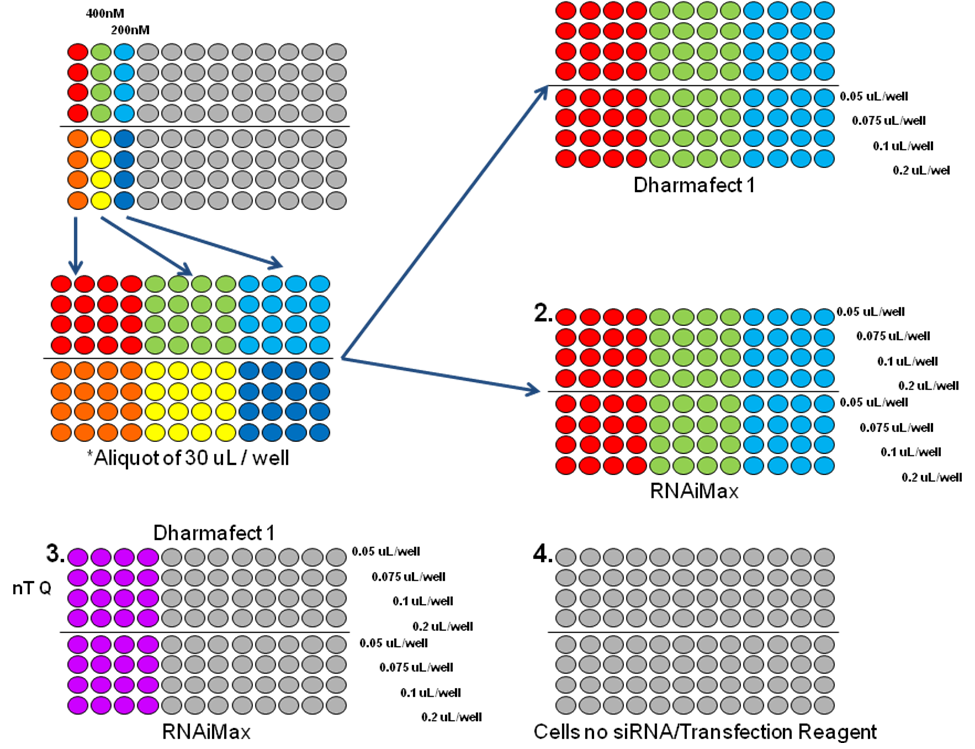
Step 2: Once the siRNA concentration, transfection reagent vendor and concentration have been established from step 1, we test the assay using those conditions to ensure the screen has met the established criteria before moving forward. When preparing to run a screen, a statistically relevant measure is needed to determine if the screen produces data with sufficient sensitivity to accurately differentiate between positive "hits" and negative samples. Three measurements are commonly used: signal-to-background ratio, coefficient of variation, and the Z' Factor. The measure we use is the Z' Factor (see below). The Optimization Step 2 procedure will include two or more plates fully loaded with positive and negative controls to fully test the outputs "robustness". The calculated Z' Factor will be a number between 0 and 1 with the best, most robust assays approaching 1.
To interpret the Z'-factor, use these guidelines:
- A Z-factor of 1, ideal. This is approached when you have a huge dynamic range with very small standard deviations.
- A Z-factor between 0.5 and 1.0 is an excellent assay.
- A Z-factor between 0 and 0.5 is marginal.
- A Z-factor less than 0 means that the signals from the positive and negative controls overlap, making the assay essentially useless for screening purposes. [1]

In addition to the Z' Factor, which will measure the "robustness" of the assay, there are several methods we can use to visualize the reproducibility of the output. These visualization techniques are utilized to identify systematic sources of error or data with poor reproducibility due to poor assay design or implementation. Finally, a Knock-down time course experiment is run to determine the Knock-down window of the test proteins. This will establish to optimal time frame in which the experiment should be run.
Raw data Heat Maps:

The above is an example of a 384 well small compound screen plate with no edge effects (Carter, Sharma, and Ebner, unpublished data)
Replicate Correlation Plot:
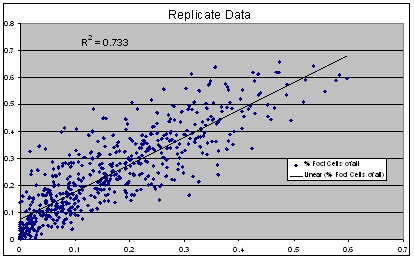
(Lunden, Ebner, unpublished data)
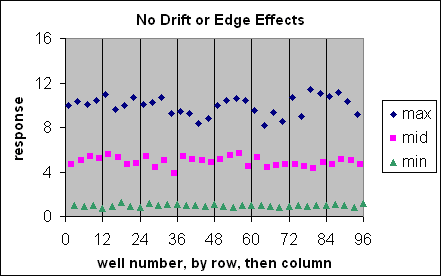
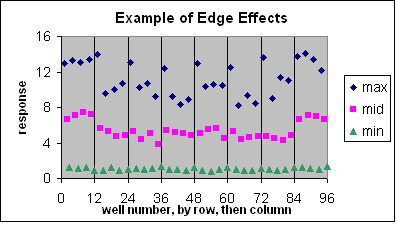
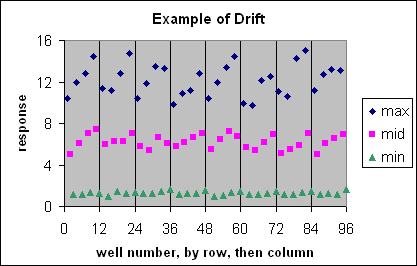
These are visual examples of Edge Effect and Drift from the Eli Lilly and Company and the National Institutes of Health Chemical Genomics Centre guidelines for running HTS assays.
Screening without a Positive Control
Not all screens have a positive control to accurately measure the phenotypic change a positive hit in your screen might produce. Often, finding those hits is the reason for running a novel screen and if there was a positive control, there would be no reason to run the screen! In these situations, extra care must be applied to develop the screen. Additionally, in siRNA screens, controls for silencing (PC-S), must be included to ensure the assay conditions produce the highest level of knock-down. In most cases, if the assay is performing well, positive hits follow.
Knock-Down Time Course:
Below is a good example of differential knock-down observed between two genes using the same transfection conditions over 72 hours.
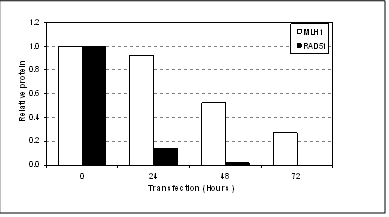
*Delay in knock-down of MLH1 (72 hours) when compared to RAD51 (24 hours). (N.Pedley, unpublished data) Reasons for the delay in siRNA knock-down can include:
- 1. High endogenous expression level of the gene
- 2. The protein products activity and half-life
RNAi Transfection Protocols - Two Approaches
There are two RNAi HTS transfection protocols used in TDI. Both are Reverse Transfection procedures but they differ in how the replicate plates are produced. Each protocol has strengths and weaknesses.
HTS RNAi Transfection Protocol Approach 1 -
A single plate of cells is transfected using a reverse transfection procedure and after 24 hours, the cells are trypsinized and split into multiple plates.
Strengths:
- Uses small amounts of siRNAs, which reduces reagent cost
- Can reduce plate to plate transfection variability because each plate is a replicate of the original transfection plate
Weaknesses:
- Time consuming because splitting cells after 24 hours requires a large amount of liquid handling
- Relies on accurate and efficient transfection in each well with no replicates able to compensate for outliers.
A very good example of this approach can be found in the mini-clonogenic screen (G.Higgins and R. Prevo, unpublished data).
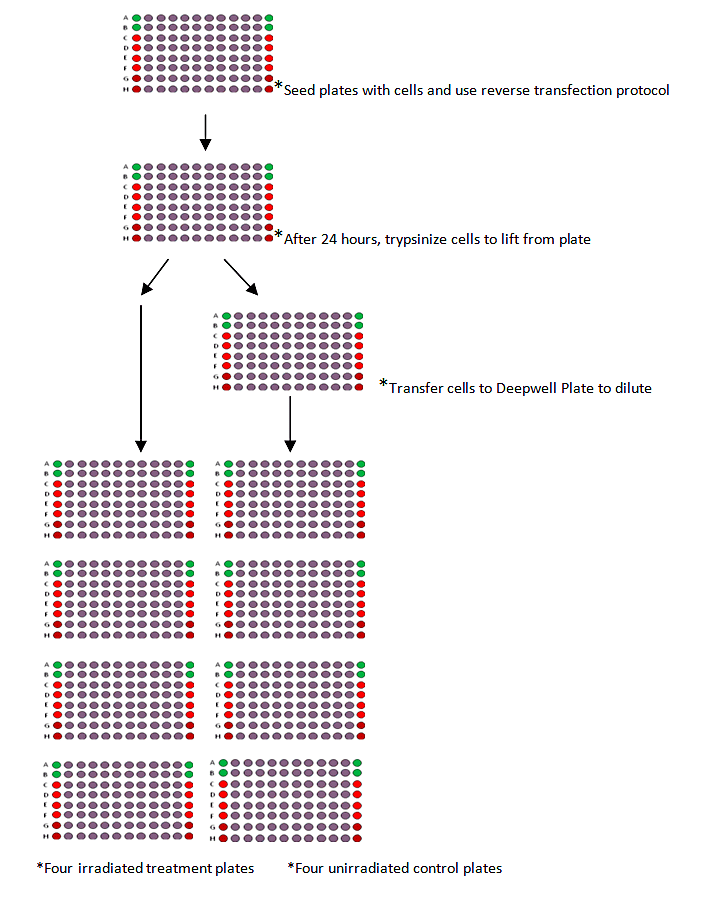
The procedure can be summarized as:
- 1. Use the reverse transfection procedure in 96 well plates to transfect each well with one siRNA.
- 2. Trypsinize the cells 24 hours later diluting the cells in a deepwell plate for the four unirradiated controls plates while the irradiated treatment plates are transferred directly from the original transfection plate.
- 3. Irradiate the treatment plates 4 hours post-transfer.
- 4. Incubate for 5-7 days.
- 5. Fix and count colonies.
A second example from the literature can be found in the Synthetic Lethal experiment using PARP inhibitor and siRNAs.[3]
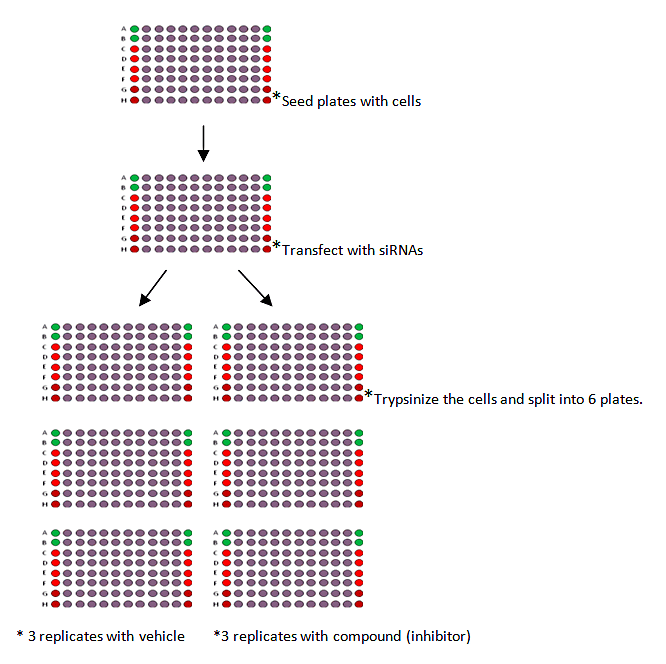
The procedure can be summerized as:
- 1. Seed cells onto 96 or 384-well plates then transfect with siRNAs 24 hours post-seeding.
- 2. Trypsinize the cells 24 hours later and split into six identical replicates.
- 3. Transfect for a total of 48 hours, remove the media and replace with 0.01% DMSO in media for the three vehicle control plates or PARP inhibitor in media (0.01% DMSO) for the three inhibitor plates.
- 4. Media containing vehicle or inhibitor was replenished after 48 hours.
- 5. Cell viability with CellTiter-Glo Luminescence Cell Viability Assay was measured after 5 days total exposure to inhibitor.
HTS RNAi Transfect ion Protocol Approach 2 -
A transfection plate is produced with enough complexes to transfect three plates of cells (three replicates) using a reverse transfection procedure and after 24 hours, the media is replaced.
Strengths:
- The transfection is completed at the beginning of the assay for all replicates and only a media change is needed at 24 hours.
- Slightly reduces the number of plates used during the procedure.
- Multiple transfection replicates able to compensate for outliers.
Weaknesses:
- Uses larger amounts of siRNAs which increases reagent cost.
- Can have larger plate to plate variability.
A very good example of this approach can be found in the RAD51 Foci Screen (C. Lundin, unpublished data)

The procedure can be summarized as:
- 1. A working stock plate of 80 µL of siRNA complexes (siRNA and transfection reagent) is produced.
- 2. 20 µL of complexes are added to 30 uL of media and 50 µL of cells are immediately added. This is reverse transfection procedure in 96 well plates to transfect plate in triplicate.
- 3. Replace the media at 24 hours.
Statistical Analysis:
A well designed, highly sensitive and robust high throughput screen requires quality control and accurate measurements. These requirements work in unison to produce a screen that yields hits that can be confirmed in secondary screens as being biologically active. Design and sensitivity has been addressed previously in this guide. Here, we will address quality control and statistical analysis of raw data.
Plate-to-Plate Variability
Because systematic error can decrease the overall performance of a screen, controls are always included on assay plates to help identify plate-to-plate variability and establish background levels of an assay. It is important in HTS to have the ability to compare all of the plates in a production run to each other. To do this, normalization of raw data is employed. Below are a couple of processing methods with a summary of their equations:


Below, the Z Score, Enhanced Z Score and B Score methods excludes the plate positive and negative control measurements and instead uses the samples themselves as controls under the assumption that the majority of samples are inactive and the data is normally distributed.


*Enhanced Z Score is more resistant to outliers (positive hits) than Z Score.

*The B Score is a variant of the Z score that uses MAD to account for plate-to-plate variability as well as a two-way median polish to minimize row and column affects.

*The 3D-B Score is a variant of the B score but applies a third dimension (stacked plates) to minimize row, column and plate affects. (B. Evers, unpublished data)
We have used several of the above tools to normalize our HTS data sets with success. It is standard practice in the HTS field to use several normalizing methods to analyse your data sets before choosing the method that works best for your screen. Additional assistance can be obtained from The Computational Biology Research Group (CBRG) which provides computing support for bioinformatics analysis at the University of Oxford:
http://www.molbiol.ox.ac.uk/CBRG_home.shtml
*A note on control placement on high throughput assay plates: Most commercial high throughput libraries are produced with the outside columns empty to accommodate the placement of assay controls. When possible, it is recommended that control columns be moved to the interior of the plate to minimize positional bias (edge effects).
Replicates
Once the assay protocol and liquid handling procedures have been optimized, the only way to minimize experimental variability further is to increase the number of replicates and then average the resulting measurements. This serves two functions. Estimates based on repeated measurements are less variable than single measurements. Secondly, additional replicates reduce the number of false positives without increasing the false negatives. Of course, additional replicates incur additional cost to a primary screen. However, in our experience, these additional cost are compensated by the reduced false negative hit rate, a stronger statistical evaluation of the results (a way to estimate variability of measurements), and reduced cost associated with "cherry-picking" when repeating primary hit results. Additionally, high throughput screens we have developed at the Target Discovery Institute where lower Z' factors were obtained were successfully developed and produced validated hits by running the assay in triplicate
We recommend a primary screen, both RNAi and small compound, be run in triplicate. Here, we define replicates as three independent repeats measured under the same experimental conditions. The additional costs are mostly incurred in the plates since in most assays; working stocks of compounds and siRNA are produced from concentrated stocks (greater storage life of concentrated stocks) and are sufficient to produce three replicates. Expensive antibodies or signal producing reagents are increased as well in triplicate primary screens but this is usually a fraction of the additional cost.
Sources of Experimental Error in HTS and Reducing Their Effects
Automation of scientific assays to the HTS platform introduces the potential for experimental and responsive variability not often observed in smaller experiments performed on the bench-top. Sources of systematic error found in HTS can be:
- 1. Temperature and evaporation gradients "edge effects"
- 2. Liquid handling malfunctions - "drift" or repeated pipetting errors
- 3. Batch processing errors
- 4. Non-homogeneous cell seeding
- 5. Differential reagent degradation over time (stability)
- 6. Solubility
- 7. Variation or gradient temperatures in the incubators
- 8. The effect of each individual siRNA on cell growth
Techniques for reducing variability in High Throughput Screening
Screens in the department of TDI use several techniques for reducing assay variability between plates and between wells. To mitigate the effects of systematic error and to assist in the troubleshooting of observed error, we have implemented a stringent regiment of:
- Implementing a liquid handling procedure that to the best of the developer's ability treats all plates exactly the same. The reagents must be added and incubated in the same order and over the same amount of time. This is the developer's responsibility.
- For cell based assays, the plates must receive the cells in good condition and at the same incubation times. For assays with several steps or more replicates, unfortunately this is very difficult since cells must be kept in suspension for a substantial length of time as the plates are processed. Often there can be two to three hours between the first plate and the last plate in a production run so this can lead to substantial variability if not controlled for.
- For all cell based assays, the condition of the cells going into the screen plates is paramount. Cells must be actively dividing and under 80% confluent. We have implemented a standardized cell seeding protocol to ensure the cells are fit for the screen. We plate a known number of cells that will be used in the screen assay into T75 or T175 flasks three days before the assay knowing they should be at the correct growth stage and number necessary to seed all of the plates needed for the run. This is a crucial control if a production run is processed over the course of days or weeks.
- When possible, inhibitors or siRNAs known to produce a phenotypic change or inhibition in the screen should be included in the "body" of the screen and on multiple plates. These known positive hits must come from the library vendor and will provide information on the assay sensitivity, library condition, precision and reproducibility, and could help diagnose any sources of experimental variability.
- Recording the location and time the plates are in the incubator (it is good practice to independently verify the incubator functioning within its proper temperature, humidity and gas settings). This is more of a trouble shooting step to identify potential sources of error as a production run is progressing.[4]
- Production runs should be completed in as short a window as possible (days or weeks) to minimizing the effect of passage number on the screen. Also, the reagents used in a production screen should all originate from the same lot to reduce lot to lot variability of your reagents.
- After the cells have been seeded, the cell plates are allowed to rest at room temperature for 20 minutes to allow the cells to settle and begin reattaching to the plate surface.[5]
- When possible, additional media changes can be used for screens of longer duration (more than three days) to minimize gas gradients affecting differential cell growth across a plate.
Pre-analysis Metrics
In large high throughput screening production, it is important to monitor the quality of the data being produced during the production run to ensure the procedures are working as expected and the data is of sufficient quality to allow statistically relevant conclusions to be made.
We employ the Z Factor (a slight variant of above Z' Factor, see above) during the assay as it assesses screen samples of screened plates as they are produced.
Additionally, strictly standardized mean difference (SSMD) can be used to assess screen data which is statistically more rigorous.[6]
Identifying Positive Hits
The main goal of any high throughput screen is to identify positive hits, that is, samples that are meaningfully differentiated from the negative controls. Once all the plates have been normalized, the final task of the primary screen is to identify these "positive hits".
There are many positive hit identification techniques available and in many cases, several are used and collated do produce a hit list ranging from strong to weak positives. These techniques include:
1. Mean of normalized data + Standard Deviation - For simplicity, this is the most heavily used positive hit scoring technique and one we use most frequently at the Target Discovery Institute. Depending on the screen, the + standard deviation can be set to -2 (2) or -3 (3).
2. Median of normalized data + MAD - A more Robust technique to identify positive hits.
Once the positive hit list has been produced, the capacity for follow-up study often dictates the number of positive hits passed on for secondary screen evaluation. For example, in the case of RNAi screens, deconvoluted siRNAs can be cost prohibitive so sometimes only the strongest or most interesting positive hits are further evaluated.
Post Assay "off-target" Filtering
Once the primary screen is completed, additional filtering will be necessary to ensure the positive hits are real and not "Off-target effects". The steps below can be used to confirm primary hits:
Step 1: All positive hits will be re-evaluated for reproducibility (Filter 2) in the same assay (repeat primary screen with positive hits) to reconfirm positive hits.
Step 2: Deconvolute the pool into the 6 siRNA (4 siRNA) duplexes. Then 4 of 6 (2 of 4) must hit to be considered "on-target".
Step 3: Third party siRNA can confirm an "on-target" hit but cannot confirm / eliminate "off-target" hits. In theory this is true but in practice, if a second vendor siRNA does not confirm a hit, it is most likely "off-target".
Step 4: TaqMan Gene Expression Assays - qrtPCR confirmation that the specific gene is being knocked-down
Step 5: Repeat the remaining hits on a second cell line (if possible). The mechanism of sensitivity or loss of function might be restricted to specific cell line. Although not specifically an "off-target" strategy, this is an excellent follow-up assay and filter aid.
Reporting Results
When reporting RNAi screen results we suggest you follow the guidelines set out by MIARE Minimum Information About an RNAi Experiment. Information can be found on the web site:
http://miare.sourceforge.net/HomePage
Vector based RNAi (short hairpin RNA /shRNAi)
shRNAi is a long term (or transient) RNAi that uses viral delivery to produce gene knockdown. shRNA can be used effectively for HTS but is not always the best choice because of a number of reasons (Class II, costly reagents, etc). It is however a very powerful technique under certain circumstances and is becoming more popular for running RNAi screens on non-dividing, primary or difficult to transfect cells.
The Broad Institute's RNAi Platform has collaboration with PerkinElmer to detail a HTS procedure for shRNAi screens using a special robotics platform and have produced a paper entitled, Automated High Throughput Transfect ion: Streamlining Lentiviral RNAi Screening Workflows on the JANUS Automated Workstation.
shRNAi can however be used in HTS to silence a specific gene of interest (often using an inducible system) followed by a standard siRNA screen to identify protein associated with the shRNAi protein. In this case, shRNAi is used as an inhibitor of a specific gene when a small compound inhibitor is not available.
Open Biosystems review of Thermo (Dharmacon) shRNAi products:
http://www.openbiosystems.com/rnai/
Cell Viability Assays
A quick review of Cell Viability assays can be found on the following web site:
http://www.rndsystems.com/product_detail_objectname_cell_viability.aspx
We use the Resazurin salts in PBS (10µg/mL) for our cell viability assays due to very low cost of the reagent and high sensitivity.
HTS Review Papers and Resources
Below are a number of papers (and Web sites) that provide excellent reviews of HTS in specific fields of research. This list is representative and is not meant to replace of literature search!
NIH Chemical Genomics Center (General guidelines for HTS)
Current Trends in HTS:
Mayr, L.M. and D. Bojanic, Novel trends in high-throughput screening. Curr Opin Pharmacol, 2009. 9(5): p. 580-8.[7]
Statistics in HTS:
Malo, N., et al., Statistical practice in high-throughput screening data analysis. Nat Biotechnol, 2006. 24(2): p. 167-75.[8]
Synthetic Lethality in Cancer Research HTS review:
Canaani, D., Methodological approaches in application of synthetic lethality screening towards anticancer therapy. Br J Cancer, 2009. 100(8): p. 1213-8.[9]
Negative selection shRNAi synthetic lethality screen:
Luo, J., et al., A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell, 2009. 137(5): p. 835-48.[10]
Cardiovascular Research HTS review:
Etzion, Y. and A.J. Muslin, The application of phenotypic high-throughput screening techniques to cardiovascular research. Trends Cardiovasc Med, 2009. 19(6): p. 207-12. [11]
Cell Based Pharmacogenomics:
Welsh, M., et al., Pharmacogenomic discovery using cell-based models. Pharmacol Rev, 2009. 61(4): p. 413-29.[12]
Small compound High Content Imaging Screen review:
Carpenter, A.E., Image-based chemical screening. Nat Chem Biol, 2007. 3(8): p. 461-5.[13]
Small compound cell-based HTS target identification:
Rix, U. and G. Superti-Furga, Target profiling of small molecules by chemical proteomics. Nat Chem Biol, 2009. 5(9): p. 616-24.[14]
Saxena, C., et al., An immuno-chemo-proteomics method for drug target deconvolution. J Proteome Res, 2008. 7(8): p. 3490-7.[15]
Bioluminescent Assays for High-Throughput Screening:
Mayr, L.M. and D. Bojanic, Novel trends in high-throughput screening. Curr Opin Pharmacol, 2009. 9(5): p. 580-8.[16]
Biosensors:
Morris, M.C., Fluorescent biosensors of intracellular targets from genetically encoded reporters to modular polypeptide pTDIes. Cell Biochem Biophys, 2010. 56(1): p. 19-37.[17]
Kurzawa, L. and M.C. Morris, Cell-cycle markers and biosensors. Chembiochem, 2010. 11(8): p. 1037-47.[18]
List of Academic Screening Facilities World-wide:
References
1. Zhang, J.H., T.D. Chung, and K.R. Oldenburg, A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen, 1999. 4(2): p. 67-73.
2. Zhang, X.D. and J.F. Heyse, Determination of sample size in genome-scale RNAi screens. Bioinformatics, 2009. 25(7): p. 841-4.
3. Lord, C.J., et al., A high-throughput RNA interference screen for DNA repair determinants of PARP inhibitor sensitivity. DNA Repair (Amst), 2008. 7(12): p. 2010-9.
4. Maddox, C.B., L. Rasmussen, and E.L. White, Adapting Cell-Based Assays to the High Throughput Screening Platform: PTDIlems Encountered and Lessons Learned. JALA Charlottesv Va, 2008. 13(3): p. 168-173.
5. Lundholt, B.K., K.M. Scudder, and L. Pagliaro, A simple technique for reducing edge effect in cell-based assays. J Biomol Screen, 2003. 8(5): p. 566-70.
6. Birmingham, A., et al., Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods, 2009. 6(8): p. 569-75.
7. Merten, C.A., Screening Europe 2010: an update about the latest technologies and applications in high-throughput screening. Expert Rev Mol Diagn, 2010. 10(5): p. 559-63.
8. Malo, N., et al., Statistical practice in high-throughput screening data analysis. Nat Biotechnol, 2006. 24(2): p. 167-75.
9. Canaani, D., Methodological approaches in application of synthetic lethality screening towards anticancer therapy. Br J Cancer, 2009. 100(8): p. 1213-8.
10. Luo, J., et al., A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell, 2009. 137(5): p. 835-48.
11. Etzion, Y. and A.J. Muslin, The application of phenotypic high-throughput screening techniques to cardiovascular research. Trends Cardiovasc Med, 2009. 19(6): p. 207-12.
12. Welsh, M., et al., Pharmacogenomic discovery using cell-based models. Pharmacol Rev, 2009. 61(4): p. 413-29.
13. Carpenter, A.E., Image-based chemical screening. Nat Chem Biol, 2007. 3(8): p. 461-5.
14. Rix, U. and G. Superti-Furga, Target profiling of small molecules by chemical proteomics. Nat Chem Biol, 2009. 5(9): p. 616-24.
15. Saxena, C., et al., An immuno-chemo-proteomics method for drug target deconvolution. J Proteome Res, 2008. 7(8): p. 3490-7.
16. Fan, F. and K.V. Wood, Bioluminescent assays for high-throughput screening. Assay Drug Dev Technol, 2007. 5(1): p. 127-36.
17. Morris, M.C., Fluorescent biosensors of intracellular targets from genetically encoded reporters to modular polypeptide pTDIes. Cell Biochem Biophys, 2010. 56(1): p. 19-37.
18. Kurzawa, L. and M.C. Morris, Cell-cycle markers and biosensors. Chembiochem, 2010. 11(8): p. 1037-47.

